One of the great challenges in modern biology is understanding how changes in amino acids that are the building blocks of proteins lead to changes in the characteristics of a living organism. This Research Infrastructure Improvement Track-2 Focused EPSCoR Collaborations (RII Track-2 FEC) award addresses this challenge by using computer simulations, mathematical modeling, and experiments to determine how amino acid changes modify the way that proteins interact with other molecules.
Hypothesis
Our central scientific hypothesis : protein biophysical models provide an efficient framework for predicting how mutations – alone, in combination, and in different environments – influence protein stability, affinity for substrates and partners, and the mappings to higher-level phenotypes.
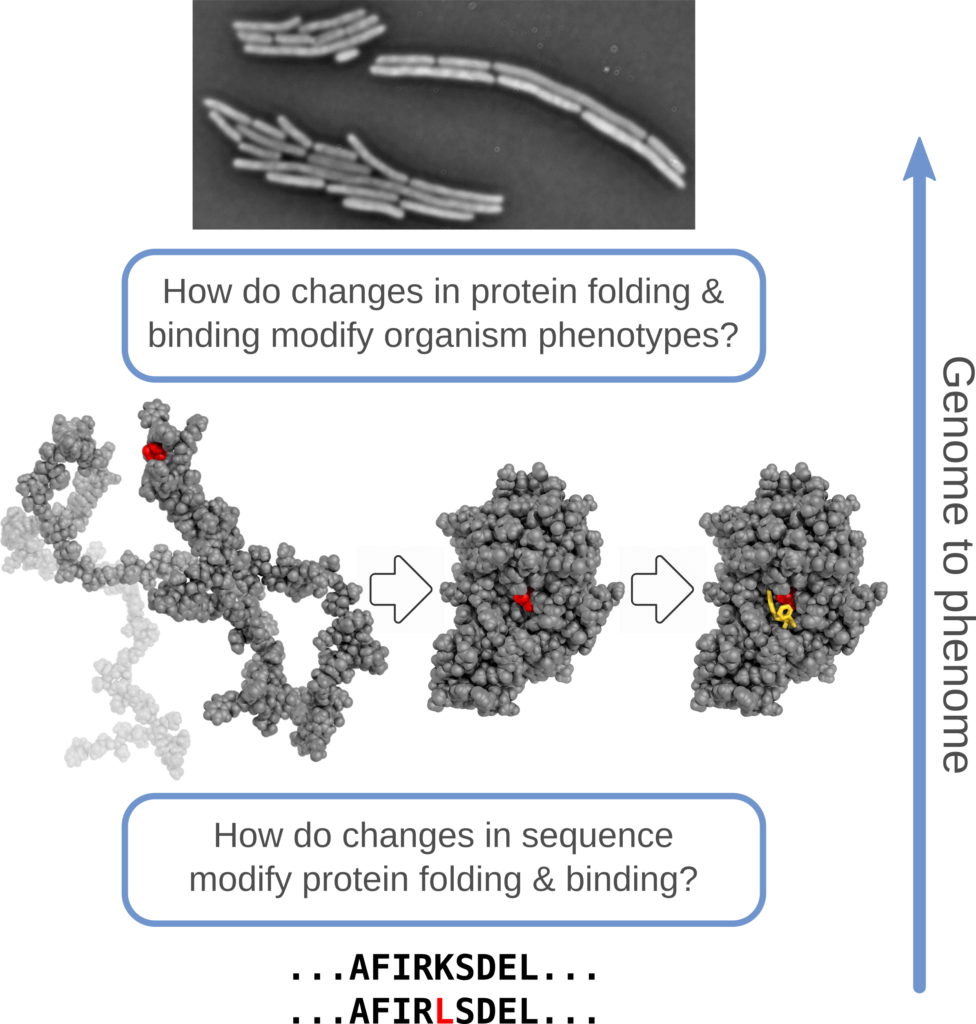
Our methodological hypothesis: this biophysics-first framework can be applied to diverse experimental systems to reveal general underlying principles of genotype to phenotype mapping, uncover gaps in understanding, and suggest potential generic and system specific extensions; these will motivate future competitive research proposals and build research capacity across all jurisdictions.
Research Objectives
We will test our scientific and methodological hypotheses via the following objectives:
Model Systems
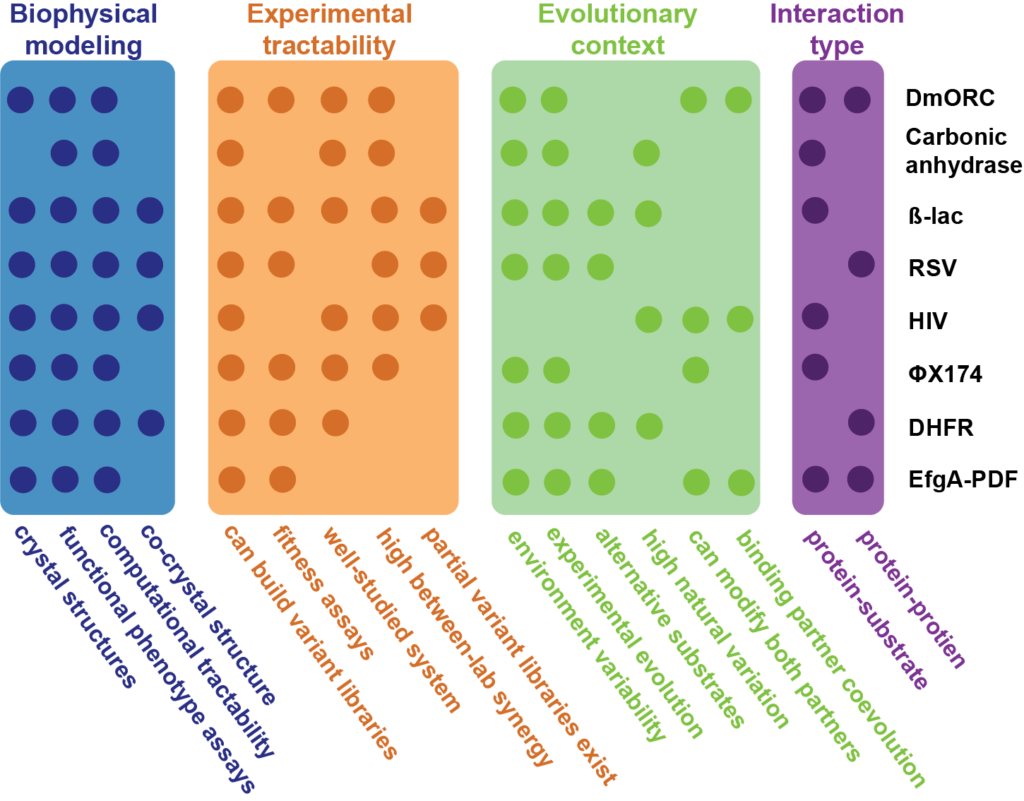
Working Groups
Our community of researchers is aggregated in “working groups,” each focused on some specific intellectual question.
GenoPheno
Biophysical characterization
Modeling
Library generation
Single-cell variation in phenotype
Diversity / inclusion
Epistasis
Outreach